Letter to the Editor – Oral microbiota, health and cancer: the dual face of the same coin in the field of the host-microbe interactome
WCRJ 2021;
8
: e2075
DOI: 10.32113/wcrj_20218_2075
Topic: Microbiota and cancer
Category: Letter to the Editor
Abstract
Dear Editor
In recent years, within the extensive research regarding human microbiota and chronic diseases, new medical and laboratory technologies have played a crucial role in understanding more complex biological pathways between human cells and microbes. On the other hand, molecular and cellular diversity and their involvement in health responses to changing host conditions have only recently been understood, through the development of brand-new laboratory tools based on ‘omics approaches. The continuous insertion of already conceptualized, new experimental large datasets and raw data onto bibliographic platforms means that they are now accessible for subsequent elaboration by other researchers. This aspect has allowed the creation of an interactive mesh of proteins, genes, and small molecules, usually called, interactome or interactomics.
Oral microbiota a “conditio sine qua non “in health and disease
The Microbiota associated with the human oral cavity represent a collective-interactive group of microbial species, mainly composed of bacteria (Firmicutes, Bacillus, Proteobacteria, and Actinomycetes) but also with some protozoa, fungi, and viral species, and is one of the most complex microbial communities in the human body1,2. These microorganisms are closely associated and disposed of as biofilm in different distinct habitats of the oral cavity, such as the tongue, teeth, mucosal surfaces, so that about 6-7 different microbiotas are present in the mouth, with the tongue dorsum showing the main extensive microbial community in the oral cavity. The types of bacteria found in oral microbiota do not change significantly; in fact, healthy people from different countries with different alimentary habits have similar compositions of oral microbiota, in this condition microorganisms coexisting in normal human tissues and provide proper functional activity in the host. This process activates the control of different fundamental processes such as immunity signal transduction, gene expression patterns and metabolism. The imbalance of oral microflora, normally called dysbiosis, has been strictly correlated with the genesis of different chronic diseases such as cancer3. For example, the anaerobic oral bacteria Fusobacterium nucleatum, which regularly lives on the tongue dorsum, normally conducts a commensal-type cell life, activating various useful biological processes such as: oral-nasal innate immunity, food flavour, and others4. In these conditions, the oral interactome network is able to modulate and maintain the commensal status of F. nucleatum. The dramatic change in microbiota bacterial composition due to a dysbiosis process is related to a significative change in the interactome network and can turn the bacterium behaviour from commensal to pathogenic5. In this case the tissue target cells are continuously stimulated through their receptors by microbiota compounds or by single bacterial proteins and injured by small molecules released in the interactome signal pathway. In the same way, target oral cells produce other related molecule-signals such as those involved in response to intensive oxidative stress (Figure 1).
Understanding the microbiological “Pandora’s box” involved in cancer
“Cancer-related bacteria” have traditionally been studied by considering their singular biochemical aspects involved in the neoplastic process function/activation or in maintaining tissue homeostasis, but this aspect lacks completeness and provides a highly limited view of the phenomenon. For example, F. nucleatum has been intensively studied in relationship to colon-rectal cancer or in some forms of oral neoplasia6. These studies often consider the bacterium as the singular biological effector in carcinogenesis, but the same approach could be observed in the literature for Porphyromonas gingivalis and with microbial genera such as: Veillonella, Actinomyces, Haemophilus, etc.7. In actual fact, this method only reveals the bacterial effector molecules for cancer, but not the complex molecular pathway due to other microbial species belonging to the microbiota or those in the target-cells molecular signals. During the tissue neoplastic transformation process and already in the first step of tumorigenesis, microbiota as well as human target cells emit a great number of molecular signals involved in: (i) the process of turning opportunistic bacteria into pathogens, (ii) chronic inflammation, (iii) anti-apoptotic molecules, (iv) related cancerogenic substances, (v) actions for evasion of the immune system8.
Some of these signals could be present years before in the oral microbiota (e.g., in the tongue biofilm) and before the tumour is clinically significant. In the same way activated cancer bacteria, through transitory bacteraemia phenomena, could move from the oral microflora to systemic tissues activating a carcinogenesis process in another body site (Figure 1).
The interactome is a way to well understand the role of oral microbiota in human health
Interactomics represents a pragmatic construct that depends on omics-based sciences. It is certainly a de novo entity that has arisen of late within the context of postgenomic systems biology.
In oral biology, this approach starts from the initial concept of protein/protein interaction between bacteria and cells, now enlarged to small molecules including metabolites and small RNAs. These molecules are normally involved in microbe-microbe and microbe-human cells communication. Freire et al9, described a singular vision regarding oral interactome networks between health and disease. In particular, these authors discuss the acquisition of a complex (mature) microbiota and adaptive immunity, as though these cell to cell relationships have evolved as a means to maintain homeostasis and where the role of the microbiota is to modulate-educate the immune system (e.g. host cytokine control) 10. These authors basically view and suppose the interactome network to be an ecological chronometer of health and disease9. If this approach is shifted to the cancer-microbe interactome and taking into consideration the heterogeneous nature of cancer, the outstanding role of this complex biological network means that it has become a new significant aspect that can contribute to the advanced diagnosis and potential treatment of cancer, particularly as regards its solid forms. The interpretation of this complex signal mesh could extend the horizon of new personalized treatments against tumors, giving a new perspective in advanced medicine that integrates clinical knowledge, bioinformatics, and networks in systems biology.
The role of Dentistry and Oral Hygiene for cancer prevention in the interactomics era
As described above, a bacteria-mediated carcinogenesis process often begins a long time before its clinical evidence. One of the first signs is represented by signals of a permanent microbial flora dysbiosis that in healthy subjects could be due to a microbial biofilm resident in the tongue dorsum or in tooth plaque. The first sign of this microbial alteration is represented by evident halitosis, due to the production of volatile sulfur compounds by bacteria11, as well as some stress-related mRNAs in the oral tissues12. This suggests that the control of dysbiosis and biofilm in oral medicine represents the first step in cancer prevention. Furthermore, the oral pathologist can promptly identify the early signs of epithelial dysplasia in the oral cavity and, by using the new high-throughput laboratory technologies, can identify the interactomics markers for diagnosis and therapy, e.g. micro and circular RNAs or other small molecules. This is a crucial aspect for efficient personalized medicine in the future. In a very immediate future, with only less than 1 minute, dentists through a simple tongue brush will can have important information for prevention and treatment of misdiagnosed cancer.
Acknowledgments
One of the authors, CC performed his/her activity in the framework of the International PhD in Innovation Sciences and Technologies at the University of Cagliari, Italy.
Conflict of Interest
The Authors declare that there is no conflict of interest.
References
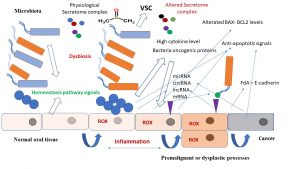
Figure 1. From dysbiosis to cancer, a schematic representation of the main interactomic signals involved in bacteria -mediated carcinogenesis in the oral cavity, for example, tongue tissue.
In recent years, within the extensive research regarding human microbiota and chronic diseases, new medical and laboratory technologies have played a crucial role in understanding more complex biological pathways between human cells and microbes. On the other hand, molecular and cellular diversity and their involvement in health responses to changing host conditions have only recently been understood, through the development of brand-new laboratory tools based on ‘omics approaches. The continuous insertion of already conceptualized, new experimental large datasets and raw data onto bibliographic platforms means that they are now accessible for subsequent elaboration by other researchers. This aspect has allowed the creation of an interactive mesh of proteins, genes, and small molecules, usually called, interactome or interactomics.
Oral microbiota a “conditio sine qua non “in health and disease
The Microbiota associated with the human oral cavity represent a collective-interactive group of microbial species, mainly composed of bacteria (Firmicutes, Bacillus, Proteobacteria, and Actinomycetes) but also with some protozoa, fungi, and viral species, and is one of the most complex microbial communities in the human body1,2. These microorganisms are closely associated and disposed of as biofilm in different distinct habitats of the oral cavity, such as the tongue, teeth, mucosal surfaces, so that about 6-7 different microbiotas are present in the mouth, with the tongue dorsum showing the main extensive microbial community in the oral cavity. The types of bacteria found in oral microbiota do not change significantly; in fact, healthy people from different countries with different alimentary habits have similar compositions of oral microbiota, in this condition microorganisms coexisting in normal human tissues and provide proper functional activity in the host. This process activates the control of different fundamental processes such as immunity signal transduction, gene expression patterns and metabolism. The imbalance of oral microflora, normally called dysbiosis, has been strictly correlated with the genesis of different chronic diseases such as cancer3. For example, the anaerobic oral bacteria Fusobacterium nucleatum, which regularly lives on the tongue dorsum, normally conducts a commensal-type cell life, activating various useful biological processes such as: oral-nasal innate immunity, food flavour, and others4. In these conditions, the oral interactome network is able to modulate and maintain the commensal status of F. nucleatum. The dramatic change in microbiota bacterial composition due to a dysbiosis process is related to a significative change in the interactome network and can turn the bacterium behaviour from commensal to pathogenic5. In this case the tissue target cells are continuously stimulated through their receptors by microbiota compounds or by single bacterial proteins and injured by small molecules released in the interactome signal pathway. In the same way, target oral cells produce other related molecule-signals such as those involved in response to intensive oxidative stress (Figure 1).
Understanding the microbiological “Pandora’s box” involved in cancer
“Cancer-related bacteria” have traditionally been studied by considering their singular biochemical aspects involved in the neoplastic process function/activation or in maintaining tissue homeostasis, but this aspect lacks completeness and provides a highly limited view of the phenomenon. For example, F. nucleatum has been intensively studied in relationship to colon-rectal cancer or in some forms of oral neoplasia6. These studies often consider the bacterium as the singular biological effector in carcinogenesis, but the same approach could be observed in the literature for Porphyromonas gingivalis and with microbial genera such as: Veillonella, Actinomyces, Haemophilus, etc.7. In actual fact, this method only reveals the bacterial effector molecules for cancer, but not the complex molecular pathway due to other microbial species belonging to the microbiota or those in the target-cells molecular signals. During the tissue neoplastic transformation process and already in the first step of tumorigenesis, microbiota as well as human target cells emit a great number of molecular signals involved in: (i) the process of turning opportunistic bacteria into pathogens, (ii) chronic inflammation, (iii) anti-apoptotic molecules, (iv) related cancerogenic substances, (v) actions for evasion of the immune system8.
Some of these signals could be present years before in the oral microbiota (e.g., in the tongue biofilm) and before the tumour is clinically significant. In the same way activated cancer bacteria, through transitory bacteraemia phenomena, could move from the oral microflora to systemic tissues activating a carcinogenesis process in another body site (Figure 1).
The interactome is a way to well understand the role of oral microbiota in human health
Interactomics represents a pragmatic construct that depends on omics-based sciences. It is certainly a de novo entity that has arisen of late within the context of postgenomic systems biology.
In oral biology, this approach starts from the initial concept of protein/protein interaction between bacteria and cells, now enlarged to small molecules including metabolites and small RNAs. These molecules are normally involved in microbe-microbe and microbe-human cells communication. Freire et al9, described a singular vision regarding oral interactome networks between health and disease. In particular, these authors discuss the acquisition of a complex (mature) microbiota and adaptive immunity, as though these cell to cell relationships have evolved as a means to maintain homeostasis and where the role of the microbiota is to modulate-educate the immune system (e.g. host cytokine control) 10. These authors basically view and suppose the interactome network to be an ecological chronometer of health and disease9. If this approach is shifted to the cancer-microbe interactome and taking into consideration the heterogeneous nature of cancer, the outstanding role of this complex biological network means that it has become a new significant aspect that can contribute to the advanced diagnosis and potential treatment of cancer, particularly as regards its solid forms. The interpretation of this complex signal mesh could extend the horizon of new personalized treatments against tumors, giving a new perspective in advanced medicine that integrates clinical knowledge, bioinformatics, and networks in systems biology.
The role of Dentistry and Oral Hygiene for cancer prevention in the interactomics era
As described above, a bacteria-mediated carcinogenesis process often begins a long time before its clinical evidence. One of the first signs is represented by signals of a permanent microbial flora dysbiosis that in healthy subjects could be due to a microbial biofilm resident in the tongue dorsum or in tooth plaque. The first sign of this microbial alteration is represented by evident halitosis, due to the production of volatile sulfur compounds by bacteria11, as well as some stress-related mRNAs in the oral tissues12. This suggests that the control of dysbiosis and biofilm in oral medicine represents the first step in cancer prevention. Furthermore, the oral pathologist can promptly identify the early signs of epithelial dysplasia in the oral cavity and, by using the new high-throughput laboratory technologies, can identify the interactomics markers for diagnosis and therapy, e.g. micro and circular RNAs or other small molecules. This is a crucial aspect for efficient personalized medicine in the future. In a very immediate future, with only less than 1 minute, dentists through a simple tongue brush will can have important information for prevention and treatment of misdiagnosed cancer.
Acknowledgments
One of the authors, CC performed his/her activity in the framework of the International PhD in Innovation Sciences and Technologies at the University of Cagliari, Italy.
Conflict of Interest
The Authors declare that there is no conflict of interest.
References
- Zhang Y, Wang X, Li H, Ni C, Du Z, Yan F. Human oral microbiota and its modulation for oral health. Biomed Pharmacother 2018; 99: 883-893.
- Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol 2018; 200: 525-540.
- Tuominen H, Rautava J. Oral microbiota and cancer development. Pathobiology 2021; 88: 116-126.
- Starkenmann C, Le Calvé B, Niclass Y, Cayeux I, Beccucci S, Troccaz M. Olfactory perception of cysteine-S-conjugates from fruits and vegetables. J Agric Food Chem 2008; 22: 575-80.
- Han YW. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol 2015; 23: 141-147.
- Dessì A, Bosco A, Pintus R, Orrù G, Fanos V. Fusobacterium nucleatum and alteration of the oral microbiome: from pregnancy to SARS-COV-2 infection. Eur Rev Med Pharmacol Sci 2021; 25: 4579-4596.
- Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat 2019; 18: 1533033819867354.
- Gholizadeh P, Eslami H, Yousefi M, Asgharzadeh M, Aghazadeh M, Kafil HS. Role of oral microbiome on oral cancers, a review. Biomed Pharmacother 2016; 84: 552-558.
- Freire M, Nelson KE, Edlund A. The oral host-microbial interactome: an ecological chronometer of health? Trends Microbiol 2021; 29: 551-561.
- Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity 2017; 46: 562-576.
- Mogilnicka I, Bogucki P, Ufnal M. Microbiota and malodor-etiology and management. Int J Mol Sci 2020; 21: 2886.
- Orrù G, Muggironi F, Mameli A, Demontis C, Arcadu B, Scano A, Denotti G, Piras V, Girometta C, Zeza B, Pilloni A. BAX gene overexpression in the tongue could warn of infection risk due to periodontal pathogens. Open Dent J 2018: 12: 1070-1078.

Figure 1. From dysbiosis to cancer, a schematic representation of the main interactomic signals involved in bacteria -mediated carcinogenesis in the oral cavity, for example, tongue tissue.
To cite this article
Letter to the Editor – Oral microbiota, health and cancer: the dual face of the same coin in the field of the host-microbe interactome
WCRJ 2021;
8
: e2075
DOI: 10.32113/wcrj_20218_2075
Publication History
Submission date: 29 Jun 2021
Revised on: 30 Jul 2021
Accepted on: 02 Aug 2021
Published online: 02 Aug 2021

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.